Design. Discover. Predict.
What is NEUROBLOX?
The platform for modeling neural control circuits and their regulation.
Douglas Rothman, Ph.D.
Botond Antal, Ph.D.
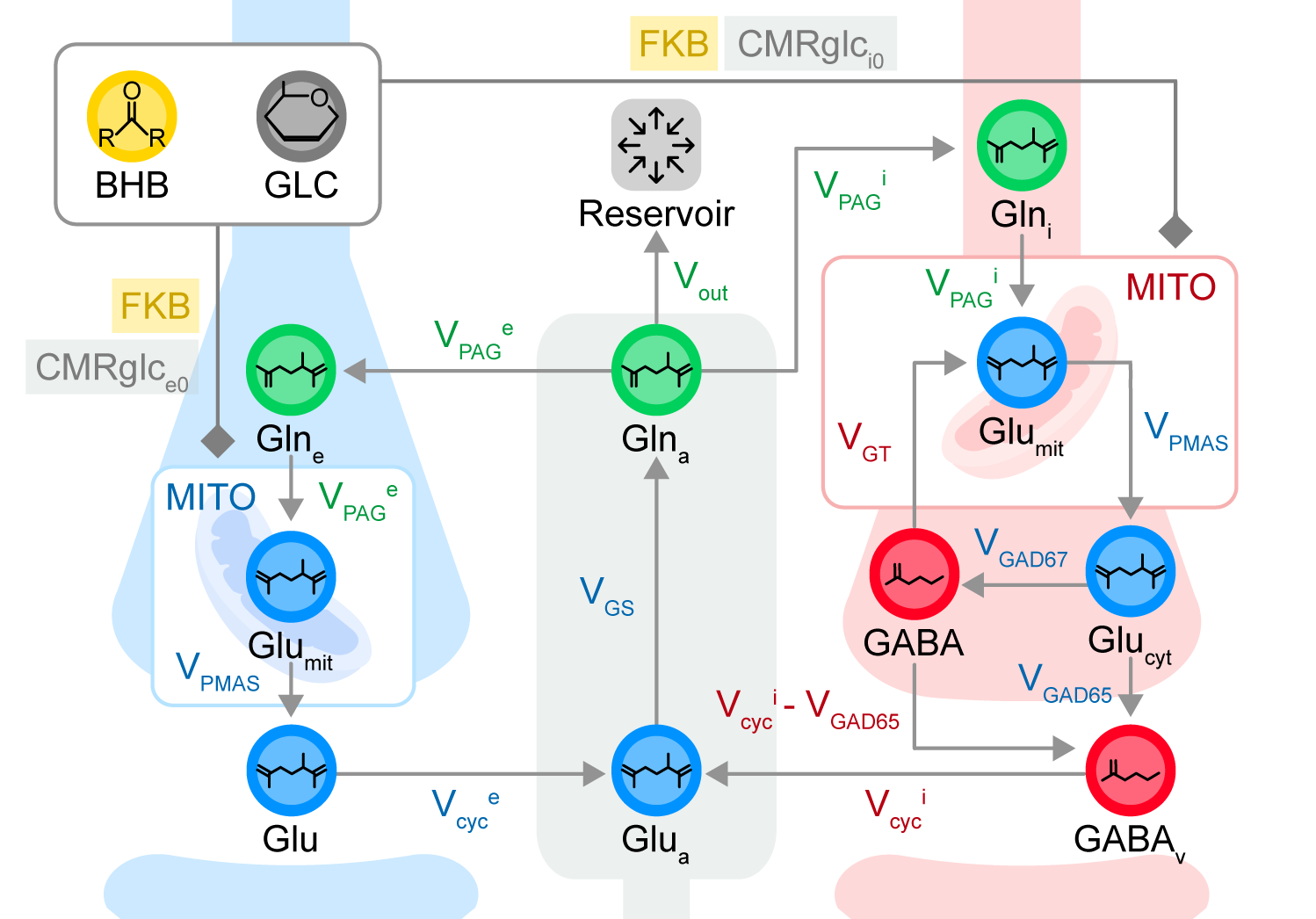
NEW Neuroblox Neurotransmitter Cycling Model
Explore how metabolic interventions like ketosis impact brain neurotransmitter levels.
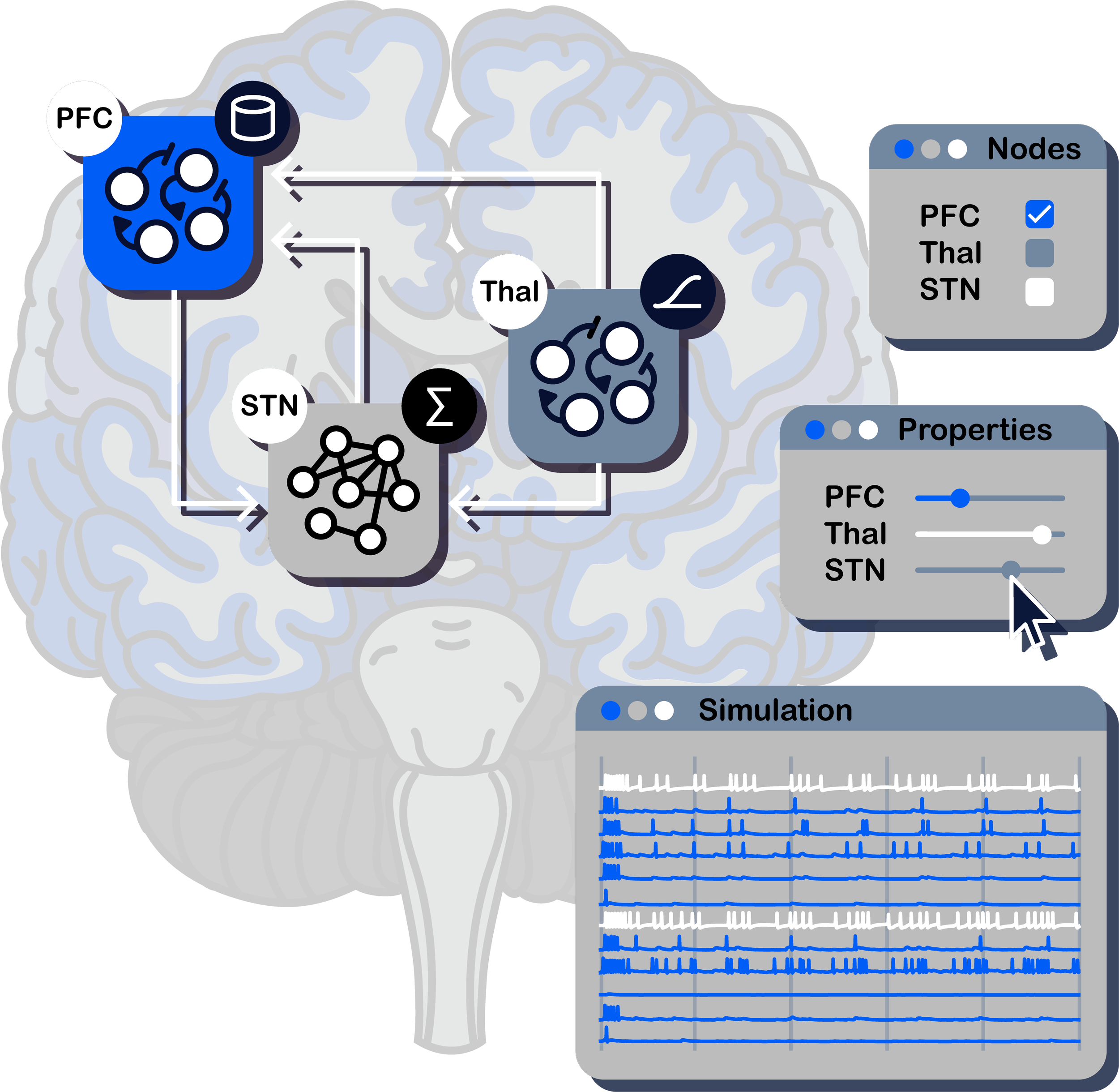
MULTISCALE.
Simulate at single-neuron to whole-brain scales.
MULTIMODAL.
Integrate diverse experimental data: patch clamp, LFP, ECOG, MEEG, fMRI, and behavior.
MULTISYSTEM.
Integration of the brain with other physiological systems. Plug-in modules for probing the impact of metabolism, pharmacological interventions, and deep brain stimulation on neural circuits and their regulation.
MODULAR.
Choose from our blocks, or build your own. Connect and combine blocks. Simulate your results.
BIOMIMETIC.
Constrain your simulations to biology with parameters resembling conditions in vivo.
INTUITIVE.
NEUROBLOX.jl’s streamlined user interface and intuitive language lower the activation energy of your investigation.
COMPARE.
Load our templates or build your own. Save your work and share with collaborators.
COMBINE.
Build bottom-up or top-down with blocks.
VALIDATE.
Mechanistically validated with intracranial electrophysiology. Clinically validated with ultra high field fMRI.
GET NEUROBLOX
Featured Publications
-
To examine the biological building blocks of thought and action, we created biologically realistic local circuits based on detailed well-cited physiological and anatomical characteristics. These biomimetic circuits were then integrated into a large-scale model of cortical-striatal interactions for category learning. The model was not trained on, but nonetheless displayed properties similar to, neurophysiological recordings from non-human primates (NHPs) performing the same task. The model had learning curves similar to the NHPs. It showed how synaptic modifications could induce changes in spiking and synchrony in the brain. The model even made novel predictions that were subsequently found in the brain, including “bad-idea neurons” that signaled impending incorrect choices after learning. This demonstrates how key computational principles can be discovered by modeling local circuitry that mimics the brain.
Read preprint in bioRXiv.
-
All fields of science depend on mathematical models. Occam's razor refers to the principle that good models should exclude parameters beyond those minimally required to describe the systems they represent. This is because redundancy can lead to incorrect estimates of model parameters from data, and thus inaccurate or ambiguous conclusions. Here, we show how deep learning can be powerfully leveraged to address Occam's razor. FixFit, our new method, uses a feedforward deep neural network with a bottleneck layer to characterize and predict the behavior of a given model from its input parameters. FixFit has three major benefits. First, it provides a metric to quantify the original model's degree of complexity. Second, it allows for the unique fitting of data. Third, it provides an unbiased way to discriminate between experimental hypotheses that add value versus those that do not. In two use cases, we demonstrate the broad applicability of this method across scientific domains. To validate the method using a known system, we apply FixFit to recover known composite parameters for the Kepler orbit model. To illustrate how the method can be applied to less well-established fields, we use it to identify parameters for a multi-scale brain model and reduce the search space for viable candidate mechanisms.
Read preprint in arXiv.
-
Using neuroimaging and electrophysiological data to infer neural parameter estimations from theoretical circuits requires solving the inverse problem. Here, we provide a new Julia language package designed to i) compose complex dynamical models in a simple and modular way with ModelingToolkit.jl, ii) implement parameter fitting based on spectral dynamic causal modeling (sDCM) using the Laplace approximation, analogous to MATLAB implementation in SPM12, and iii) leverage Julia’s unique strengths to increase accuracy and speed by employing Automatic Differentiation during the fitting procedure. To illustrate the utility of our flexible modular approach, we provide a method to improve correction for fMRI scanner field strengths (1.5T, 3T, 7T) when fitting models to real data.
Read preprint in bioRXiv. -
As a field, control systems engineering has developed quantitative methods to characterize the regulation of systems or processes, whose functioning is ubiquitous within synthetic systems. In this context, a control circuit is objectively “well regulated” when discrepancy between desired and achieved output trajectories is minimized and “robust” to the degree that it can regulate well in response to a wide range of stimuli. Most psychiatric disorders are assumed to reflect dysregulation of brain circuits. Yet, probing circuit regulation requires fundamentally different analytic strategies than the correlations relied upon for analyses of connectivity and their resultant networks. Here, we demonstrate how well-established methods for system identification in control systems engineering may be applied to functional magnetic resonance imaging (fMRI) data to extract generative computational models of human brain circuits. As required for clinical neurodiagnostics, we show these models to be extractable even at the level of the single subject. Control parameters provide two quantitative measures of direct relevance for psychiatric disorders: a circuit’s sensitivity to external perturbation and its dysregulation.
Read Article in Computational Brain & Behavior.
Support from